The Horace-SpinW interface
Defining models in Horace: a recap
Horace accepts a variety of functions to model data:
y = fn(x1, x2, ..., xn, pars) | Functions operating directly on data coordinates (e.g. gaussian peaks) |
s = fn(qh, qk, ql, en, pars) | Model $S(\mathbf{q},\omega)$ functions evaluated for each $\omega$ |
[w, s] = fn(qh, qk, ql, pars) | General model $S(\mathbf{q},\omega)$ functions |
In all cases, immediately following the coordinates, Horace expects a vector of parameter values to be fitted
After this parameter values, Horace also accepts any other input variables as model constants which will be passed to the model
The fit functions generally only accept the s = fn(qh, qk, ql, en, pars) form for $S(\mathbf{q}, \omega)$ models,
so energy convolution needs to be done by the modelling code
The SpinW spinwave method: a recap
In order to calculate the spin wave spectrum in SpinW, something like the following needs to be used:
spec = sw_obj.spinwave(hkl, 'hermit', false, 'formfact', true);
spec = sw_egrid(spec, 'component', 'Sperp', 'Evect', 0:0.05:10);
Comparing with what Horace needs, we notice that:
- The model (fittable) parameters are not set here, but much earlier in the definition of the model
- We need the combination of both
spinwaveandsw_egridto get a function of the forms = fn(qh, qk, ql, en, pars)which Horace needs
Fortunately the wrapped model function is provided in SpinW: the method spinw.horace_sqw
The spinw.horace_sqw method
horace_sqw has the same signature as a standard Horace $S(\mathbf{q}, \omega)$ function,
horace_sqw(qh, qk, ql, en, pars, varargin)
So, it can be used directly in a Horace multifit_sqw call.
In order to define which model parameter is to be varied in the fit, you have to give horace_sqw
a mat parameter which is a cell array of the matrix names to be varied in the order they appear in
the pars vector
Since the parameters of pars are scalars, if the matrix you refer to is not isotropic (e.g. it's not
representing a Heisenberg interaction), a special syntax to refer to which matrix element(s) needs to vary has to be used.
A simple example:
J = 1.2;
K = 0.1;
tri = sw_model('triAF', J);
tri.addmatrix('label', 'K', 'value', diag([0 0 K]));
tri.addaniso('K');
fwhm = 0.75;
scalefactor = 1;
ws = cut_sqw(sqw_file, [0.05], [-0.1, 0.1], [-0.1, 0.1], [0.5]);
fitobj = multifit_sqw(ws);
fitobj.set_fun(@tri.horace_sqw);
fitobj.set_pin({[J K fwhm scalefactor], 'mat', {'J_1', 'K(3,3)'}, ...
'hermit', false, 'useFast', true, 'formfact', true});
ws_sim = fitobj.simulate();
[ws_fit, fit_dat] = fitobj.fit()
The vector [J K fwhm scalefactor] is the parameters vector. We need to tell SpinW that it corresponds to the
Heisenberg nearest neighbour interaction J_1 and the easy-place anisotropy K
Because J is isotropic, we can just give the matrix name in mat
But, K only applies to the zz element, so we need to tell SpinW that in mat
fwhm and scalefactor are parameters which are added by horace_sqw
to denote the energy FWHM and intensity scale factor (may be omitted, in which case it is taken to be unity and fixed)
The other (non-varying) parameters we pass to multifit are just standard SpinW keyword arguments
There are a few keyword arguments unique to horace_sqw
-
'useFast'- This tellshorace_sqwto use a faster but slightly less accurate code thanspinwave.In particular, this code achieves a speed gain by: - Only calculating
Sperprather than full $S^{\alpha\beta}$ tensor - Only calculating magnon creation (positive energy / neutron energy loss) modes.
- Ignoring twins
-
'partrans'- A function handle to transform the input parameters received from Horace before passing to SpinW -
'coordtrans'- A $4\times 4$ matrix to transform the input $(Q_h, Q_k, Q_l, \hbar\omega)$ coordinates received from Horace before passing to SpinW -
'resfun'- This is tellshorace_sqwwhat function to use for the energy convolution. Options are: -
'gauss'- a gaussian (one parameter: fwhm) -
'lor'- a lorentzian (one parameter: fwhm) -
'voigt'- a pseudovoigt (two parameters: fwhm and lorentzian fraction) -
'sho'- a damped harmonic oscilator (parameters: Gamma Temperature Amplitude) - A function handle to a function which will be accepted by Horace's
disp2sqwmethod
horace_sqw appends the parameters needed by resfun to the end of the parameter
vector and then adds a scale factor between the data and calculation after that
The 'mat' argument
Horace expects a parameter vector, so we have to tell SpinW which parameter is which
In simple cases, just the name of the corresponding SpinW matrix, or a string denoting which single matrix element suffice
For more complicated cases, an additional parameter 'selector', a $3\times3$ logical matrix needs to be used
This tells the matparser function which SpinW uses to decode the 'mat' argument which matrix
elements the parameter corresponds to
Dvec = [0.1 0.2 0.3];
swobj.addmatrix('label', 'DM', 'value', Dvec);
swobj.addcoupling('mat', 'DM', 'bond', 1);
sel(:,:,1) = [0 0 0; 0 0 1; 0 -1 0]; % Dx
sel(:,:,2) = [0 0 1; 0 0 0; -1 0 0]; % Dy
sel(:,:,3) = [0 1 0; -1 0 0; 0 0 0]; % Dz
fitobj.set_fun(@swobj.horace_sqw);
fitobj.set_pin({Dvec, 'mat', {'DM', 'DM', 'DM'}, ...
'selector', sel, 'hermit', false})
fitobj.fit() 'selector' is a $3\times 3\times N$ array where $N$ is the number of parameters
Each $3\times 3$ matrix denotes which elements of the corresponding matrix in 'mat' goes with that parameter
Mex files in SpinW
SpinW mex files
SpinW contains a few C libraries to speed up the spin wave calculations
They perform standard linear algebra algorithms on stacks of matrices in parallel.
eig_omp | Calculates eigenvalues / eigenvectors of a stack of matrices |
chol_omp | Calculates the Cholesky factorisation of a stack of matrices |
sw_mtimesx | Calculates matrix-matrix or matrix-vector multiplications |
For each $\mathbf{q}$ SpinW has to diagonalise or solve a Hamiltonian matrix. If this could be done in parallel there will be a speed up (typically around 2-3 times for a 4-core computer).
The 'optmem' option in spinwave determines how many $\mathbf{q}$'s are calculated
at once - the more $\mathbf{q}$'s, the greater the speed up (hence more memory is better!)
Using mex files in SpinW
To tell SpinW to use the mex'ed libraries, do:
swpref.setpref('usemex',true)
You can just type swpref again to show all the options
If the distributed compiled mex files are not compatible with your system you will get an error
Compiling SpinW mex files
The SpinW distribution contains compiled versions of these files for the standard 64-bit operating systems (Windows, Linux and Mac OS X), but in case you need to compile it yourself...
First, to compile the mex files, you will need to have installed a Matlab-compatible compiler - see: https://uk.mathworks.com/support/compilers.html
This is generally Visual Studio in Windows, gcc on Linux and XCode on Mac.
(Note that on Mac OS X you will need to use homebrew as OpenMP is not supported by the standard XCode compiler)
Set up mex using:
mex -setup
Then compile the SpinW mex files with:
sw_mex
Or:
sw_mex('test', true)
To run a test suite which will also tell you how much of a speed up you might expect (it takes ~5min to run).
Example of Horace-SpinW integration
Download the scripts here:
pcsmo_eval.m
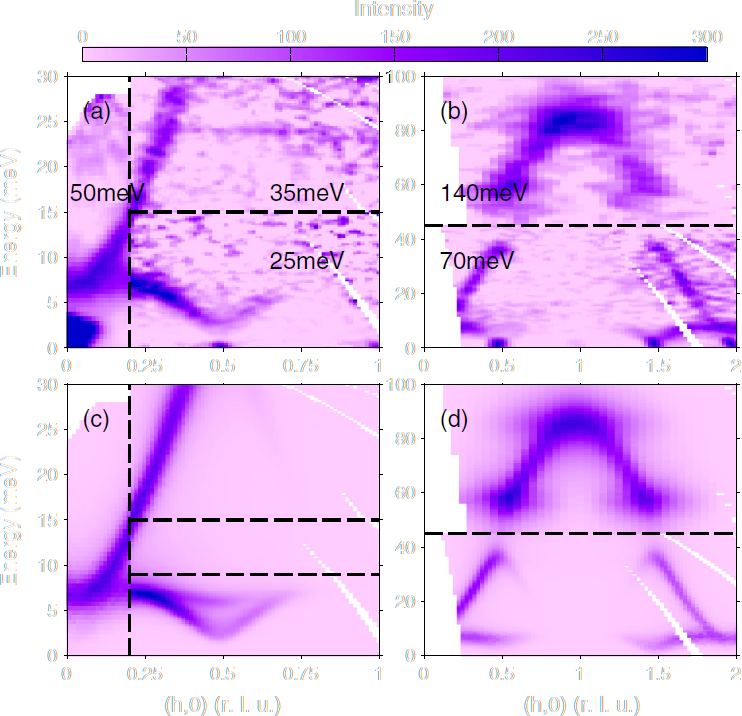
Pr(Ca0.9Sr0.1)2Mn2O7 Horace data
The .sqw files should be in the folders previously used for Horace
If you're not using a desktop, you can get the files from: Dropbox
Make 2D slices along $(h00)$ and energy transfer and compare to figure 2 of Johnstone et al.
ei = [25 35 50 70 140];
proj = projaxes([1 0 0], [0 1 0]);
for ii = 1:numel(ei);
sqw_file = sprintf('../aaa_my_work/pcsmo_ei%d_base.sqw', ei(ii));
ws_cut(ii) = cut_sqw(sqw_file, proj, [-5,0.025,5], [-0.2,0.2], ...
[-inf,inf], [ei(ii)/100]);
% Symmetrise about h=0
ws_cut(ii) = symmetrise_sqw(ws_cut(ii), [0 0 1], [0 1 0], [0 0 0]);
plot(ws_cut(ii))
lz 0 10
keep_figure;
end
Background subtraction
As in the Horace tutorials, make a cut at high $q$ and use replicate to generate a 2D background slice
Subtract this from the data and compare it to the paper
for ii = 1:numel(ei)
idx = find(sum(ws_cut(ii).data.s,2)>0);
qmax = ws_cut(ii).data.p{1}(idx(end)) * 0.5;
ws_bkg(ii) = cut_sqw(ws_cut(ii), [qmax-0.1,qmax], []);
ws_sub(ii) = ws_cut(ii) - replicate(ws_bkg(ii), ws_cut(ii));
plot(ws_sub(ii))
lz 0 10
lx -2 2
keep_figure;
end
Evaluate the SpinW model
Use the code from the previous ("real world") tutorial to set up a SpinW model of the Goodenough model
Evaluate this model on the cuts you've made:
- Because the model had to use a larger ($2\times 2\times 1$) "structural" unit cell, we need to convert the $Q_h$ and $Q_k$ coordinates given by Horace (multiply them by 2) to get the SpinW equivalent:
- Add the usual SpinW options:
cpars = {'coordtrans', diag([2 2 1 1])}
cpars = {cpars{:}, 'mat', {'JF1', 'JA', 'JF2', 'JF3', 'Jperp', 'D(3,3)'}, ...
'hermit', false, 'optmem', 0, 'useFast', true, 'formfact', true, ...
'resfun', 'gauss'};
Select the 170 meV dataset to see the overal dispersion (including gap)
Then set up a multifit object on this dataset - use a dnd object to save time
(the sqw object will have ~100x the number of pixels)
Finally tell SpinW to use mex files to speed up the calculation and evaluate the SpinW model
idx = 5; % EIs: [25 35 50 70 140], index 5 == 140meV
fwhm = ei(idx)/30; % Typical resolution ~ 3% of Ei
kk = multifit_sqw(d2d(ws_sub(idx)));
kk = kk.set_fun (@pcsmo.horace_sqw, {[JF1 JA JF2 JF3 Jperp D fwhm] cpars{:}});
kk = kk.set_free ([1, 1, 1, 1, 1, 1, 1]);
kk = kk.set_options ('list',2);
swpref.setpref('usemex',true);
% Time a single iteration
tic
wsim = kk.simulate;
t_spinw_single = toc;
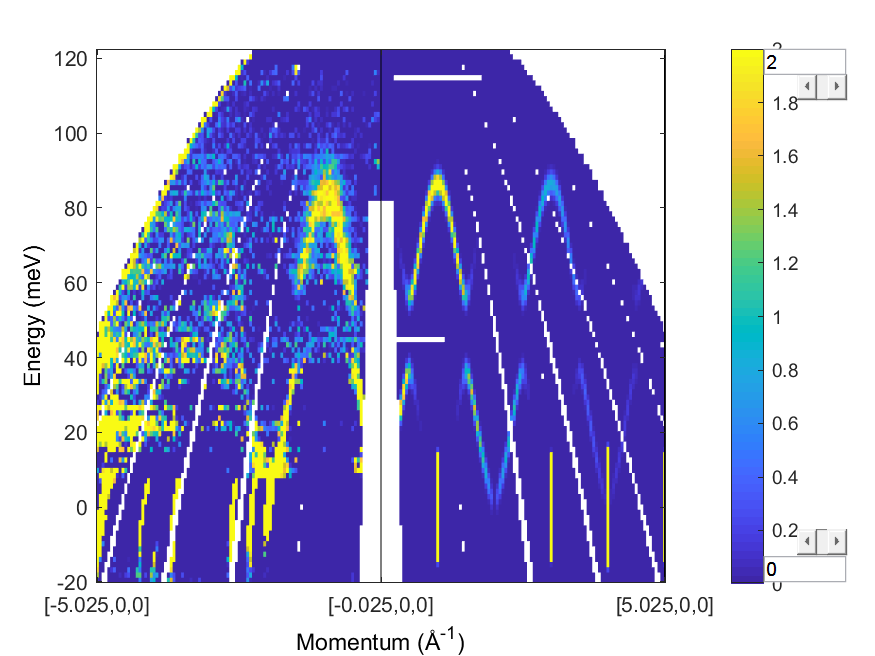
Plot the simulation
Reflect the data back to the negative $Q_h$ side, remove the empty bins (with compact )
Convert the data to a d2d object and then use spaghetti_plot to plot the
data and simulation (already a d2d object) together
wss = symmetrise_sqw(ws_sub(idx),[0 1 0],[0 0 1],[0 0 0]);
spaghetti_plot([compact(d2d(wss)) compact(wsim)])
lz 0 2
Example of Horace-SpinW fitting
Download the scripts here:
fe_fit.m
Make the set of standard $Q_h$ 1D cuts at different energies as before
proj = projaxes([1 1 0], [-1 1 0]);
energy_range = [80:20:160];
for i = 1:numel(energy_range)
my_cuts(i) = cut_sqw(sqw_file, proj, [-3,0.05,3], [-1.05,-0.95], [-0.05,0.05], ...
[-10 10]+energy_range(i));
end
Run the same fits with the analytical $S(\mathbf{q},\omega)$ function as before and note the fitted parameters and how long it takes
Define a SpinW model for bcc-Fe with a single nearest neighbour exchange and a small easy axis anisotropy
a = 2.87;
fe = spinw;
fe.genlattice('lat_const', [a a a], 'angled', [90 90 90], 'spgr', 'I m -3 m') % bcc Fe
fe.addatom('label', 'MFe3', 'r', [0 0 0], 'S', 5/2, 'color', 'gold')
fe.gencoupling()
fe.addmatrix('label', 'J1', 'value', 1, 'color', 'gray')
fe.addmatrix('label', 'D', 'value', diag([0 0 -1]), 'color', 'green')
fe.addcoupling('mat', 'J1', 'bond', 1)
fe.addaniso('D')
fe.genmagstr('mode', 'direct', 'S', [0 0 1; 0 0 1]'); % Ferromagnetic
plot(fe, 'range', [2 2 2])
Set the parameters for the fits, and also SpinW options
Note that the analytical expression used previous for $S(\mathbf{q},\omega)$ used $JS$, whereas SpinW uses $J$, and we have defined $S=\frac{5}{2}$
% Initial parameters:
J = -16;
D = -0.1;
gam = 66;
temp = 10;
amp = 131;
cpars = {'mat', {'J1', 'D(3,3)'}, 'hermit', false, 'optmem', 1, ...
'useFast', true, 'resfun', 'sho', 'formfact', true};
swpref.setpref('usemex',true);
Notice that we use the sho resolution function, in common with
the analytical expressions
Because the cuts are small, we also force SpinW to calculate all q-points in
one chunk with the {'optmem', 1} option.
Set up the multifit object on the array of 1D cuts
Use the horace_sqw method as the fit function and parameters
as defined previously, and run the fit
kk = multifit_sqw (my_cuts);
kk = kk.set_fun (@fe.horace_sqw, {[J D gam temp amp] cpars{:}});
kk = kk.set_free ([1, 0, 1, 0, 1]);
kk = kk.set_bfun (@linear_bg, [0.1,0]);
kk = kk.set_bfree ([1,0]);
kk = kk.set_options ('list',2);
[wfit, fitdata] = kk.fit('comp');
Compare the fits - are the parameters or correlation matrix consistent?