Syntax
addaniso(obj, matrixIdx, {atomTypeIdx}, {atomIdx})
Description
addaniso(obj, matrixIdx, {atomTypeIdx}, {atomIdx}) assigns the
\([3\times 3]\) matrix selected by matrixIdx (using either the matrix
label or matrix index) to the magnetic sites selected by atomTypeIdx
that can contain a name of an atom or its atom index (see spinw.atom).
If atomTypeIdx is not defined, anisotropy will be assigned to all
magnetic atoms.
Examples
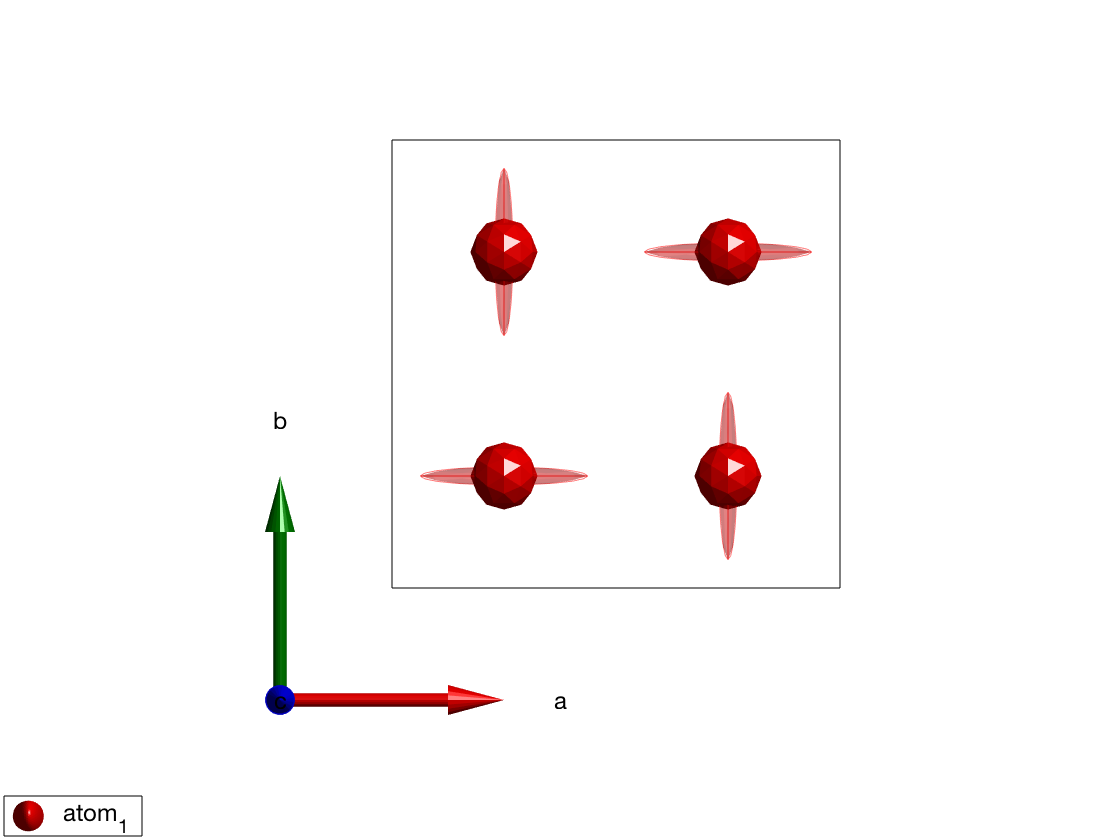
To show the effect of a fourfold axis on anisotropy, we add \(A_1\) easy-axis anisotropy to atoms at \((1/4,1/4,1/2)\) and plot the result. The 3D plot shows anistropy using ellipsoid around the magnetic atoms.
cryst = spinw
cryst.genlattice('lat_const',[4 4 3],'spgr','P 4')
cryst.addatom('r',[1/4 1/4 1/2],'S',1)
cryst.addmatrix('label','A1','value',diag([-0.1 0 0]))
cryst.gencoupling
cryst.addaniso('A1')
plot(cryst)
Input arguments
obj- spinw object.
Name-Value Pair Arguments
matrixIdx- Either an integer, that selects the matrix according to
obj.matrix.mat(:,:,matrixIdx), or a string identical to one of the previously defined matrix labels, stored inobj.matrix.label. atomTypeIdx- String or cell of strings that select magnetic atoms by
their label. Also can be a vector that contains integers, the index of
the magnetic atoms in
obj.unit_cell, with all symmetry equivalent atoms. Maximum value is \(n_{atom}\), if undefined anisotropy is assigned to all magnetic atoms. Optional. atomIdx- A vector that contains indices selecting some of the
symmetry equivalent atoms. Maximum value is the number of symmetry
equivalent atoms generated corresponding to
atomTypeIdxsite. If crystal symmetry is not 0,atomIdxis not allowed, since the anisotropy matrix for equivalent atoms will be calculated using the symmetry operators of the space group. Optional.
Output Arguments
The function adds extra entries in the obj.single_ion.aniso field of the
obj spinw object.
See Also
spinw, spinw.single_ion, spinw.addcoupling, spinw.addg and spinw.addmatrix